Biomedical Image Analysis
Biomedical Image Analysis
Welcome
to the Oxford Biomedical Image Analysis (BioMedIA) cluster
The Oxford Biomedical Image Analysis (BioMedIA) cluster is an academic group of faculty, postdoctoral researchers, software engineers, support staff and research students that develop medical imaging and image analysis algorithms and tools that aim to improve image-based diagnostics, therapies and monitoring technologies in hospitals and primary care, and for both western world and global health care settings. The breadth of our interests span all major clinical imaging modalities (particularly magnetic resonance imaging, ultrasound imaging, endoscopy imaging, histopathology), multi-modal imaging (imaging and audio, imaging and gaze tracking, imaging and electrocardiogram) and microscopy.
We conduct inter-disciplinary translational research with clinical partners in Oxford and elsewhere in the UK and overseas in clinical domains of application ranging from fetal development, to oncology, respiratory medicine, gastroenterology, neurology and cardiovascular medicine.
We are well-connected within Oxford with other major research initiatives such as the Oxford Biomedical Research Centre and the CRUK Oxford Centre.
We have a strong tradition of postgraduate research training and welcome enquiries from prospective research students. Current research students are enrolled on one of a number of programmes including: the DPhil in Engineering Science, the Centre for Doctoral Training in Health Data Sciences, and the National Institutes of Health Oxford-Cambridge Scholars Programme.
Oxford BioMedIA is part of the Institute of Biomedical Engineering in the Oxford University, Department of Engineering Science.
Our Research
To find out more about our research visit the individual group pages below.
You can also view our publications here.
Bulte Group
We work on creating image acquisition and analysis methods for obtaining quantitative physiological information from both healthy and diseased tissue. We are particularly interested in imaging vascular pathologies, hypoxia in tumours, and neurodegeneration. Our research incorporates a variety of MRI techniques including Oxygen Enhanced MRI, arterial spin labelling, T1, T2 and T2 mapping, and diffusion imaging. In addition, we have ongoing research using transcranial Doppler ultrasound, near infrared spectroscopy, pupillometry, PET and CT. The group is working towards the clinical translation of these emerging methods, and thus we concentrate on using short, patient-friendly paradigms.
As a group, we also have a strong interest in women’s health. Our research in this area focuses on pregnancy-related diseases and complications, breast cancer in younger (pre-menopausal) women, and the risk factors associated with breast cancer development. The overarching goal is to improve diagnosis and disease monitoring through advanced imaging techniques, while also building broader awareness and understanding of women’s health. In addition to our research, the group leads the IBME Women’s Health Book Club, fostering an open and collaborative environment to discuss women’s health, data collection, and related topics, aiming to raise awareness, strengthen understanding, and build unity in this important area.
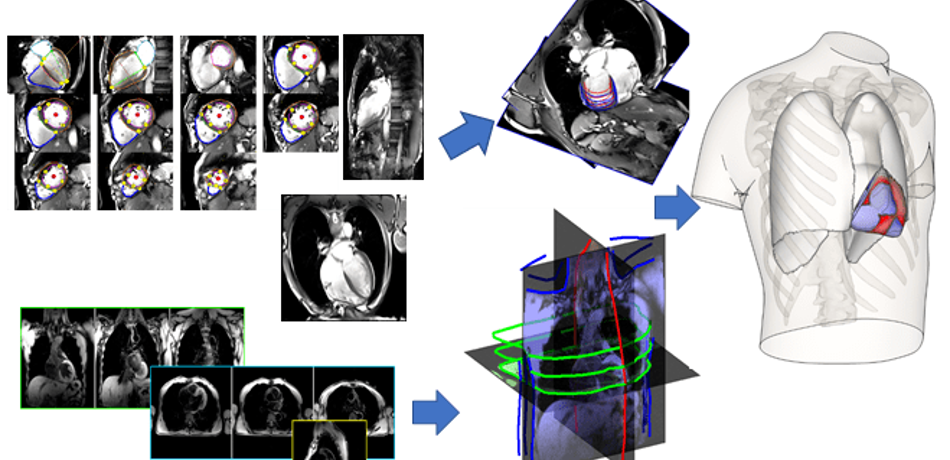
Grau Group
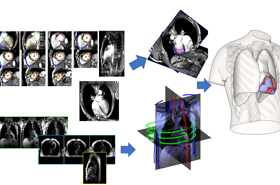
Our research focuses on the development of biomedical image analysis algorithms, with an emphasis on the combination with computational models, and applications on cardiac and pulmonary medicine.

Kamnitsas Group
We conduct advanced research on Machine Learning (ML) and Artificial Intelligence (AI) to develop the next generation of methods for medical image analysis. We push the boundaries of this technology to create more accurate, reliable and explainable methods. Our aim is to enable safe and effective use of ML/AI in healthcare, and apply this technology to tackle challenging clinical questions towards improving diagnosis and treatment.
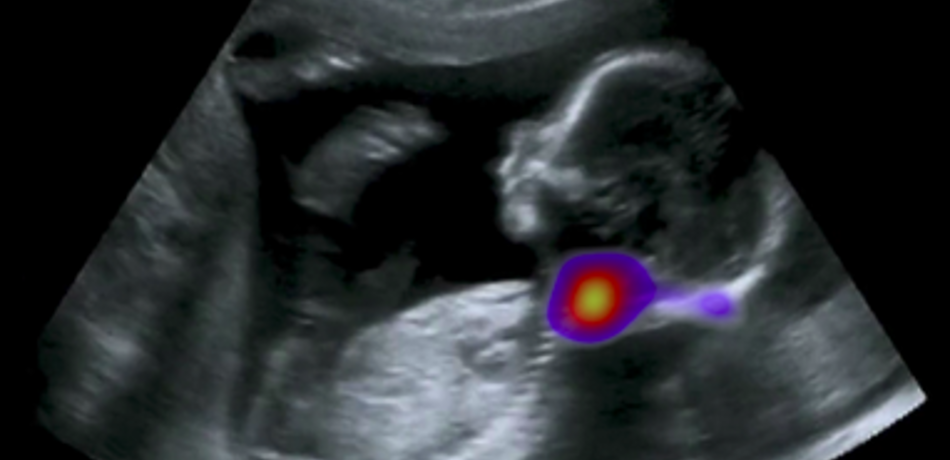
Noble Group
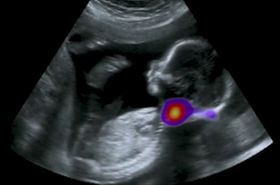
The Noble group conducts research on machine learning applied to fetal ultrasound image analysis, cardiac magnetic resonance image analysis, and cell image analysis. A major focus of the group’s research is ultrasound. We are measuring at scale how operators scan, by recording eye and probe tracking, ultrasound video and audio in the clinic. This enables us to develop machine learning models to characterise operator skill, and to build multi-modal ultrasound assistive technologies to support non-experts to scan. We are also developing low-cost ultrasound technologies empowered by deep learning based algorithms for pregnancy risk assessment in collaboration with partners in Africa and India. Our overall ambition is to advance ultrasound towards it becoming an accessible medical imaging technology to all medical professionals worldwide.
Papiez Group
Motion is a massive barrier in cancer imaging hampering developments in cancer research and treatment. My research work has comprised the development of accurate, thus complex and realistic but still computationally efficient, models of organ motion, and has established a solid foundation to reliable quantitative cancer image analysis.

Rittscher Group
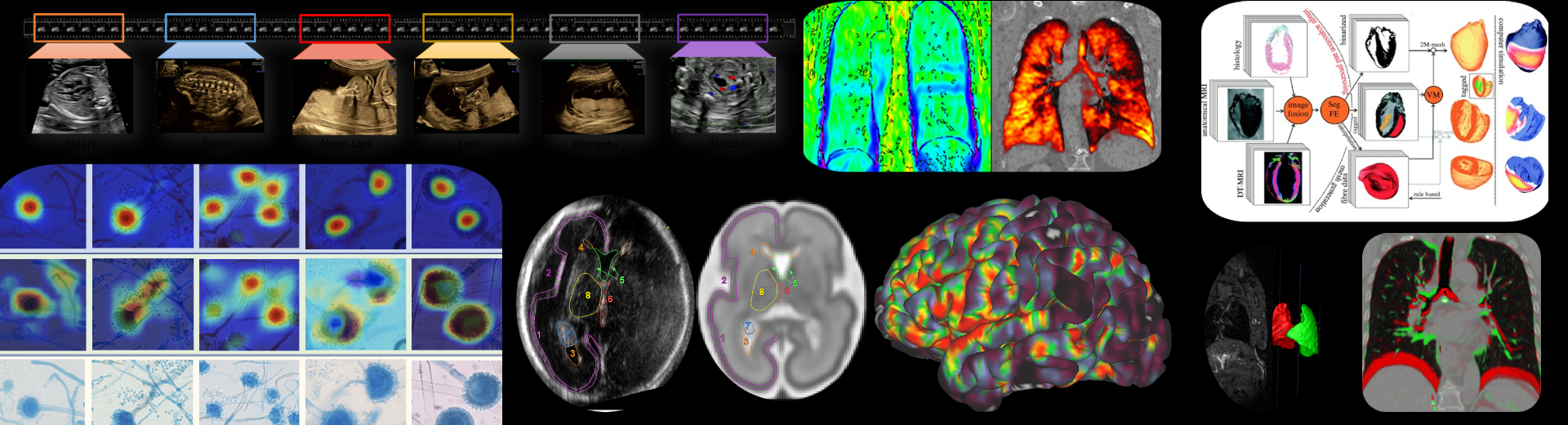
The aim of our research is to enhance our understanding of complex biological processes through the analysis of image data that has been acquired at the microscopic scale. We develops algorithms and methods that enable the quantification of a broad range of phenotypical alterations, the precise localisation of signalling events, and the ability to correlate such events in the context of the biological specimen.