03 Mar 2023
Efficient language models could help identify and understand complex medical conditions more accurately and quickly
Highly efficient and accurate models which can perform biomedical tasks have been developed by an Oxford team and made available as open source for use by healthcare professionals and biomedical researchers
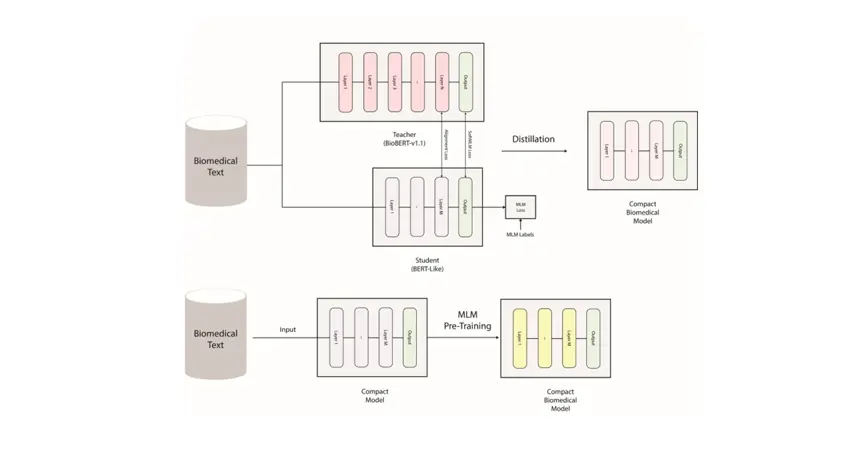
The two general strategies proposed for training compact biomedical models
Clinicians and researchers are increasingly required to analyse large amounts of data, a time consuming and labour-intensive task. In the biomedical field, tools developed from the field of natural language processing (NLP) are being applied to gain meaning from repeated sequences within large data sets. Language models can help researchers and healthcare professionals to identify and understand complex medical conditions with greater accuracy and speed.
A team at the CHI lab in the Institute of Biomedical Engineering (IBME), headed by Professor David Clifton, is developing efficient specialised language models which are much smaller and faster than existing biomedical models and can help researchers and organisations in the biomedical field to do research. They are making all the models publicly available and open source to contribute to the advancement of biomedical research and help researchers and organisations that do not have access to the latest computing infrastructure.
"Our project focuses on developing compact and efficient models that can perform biomedical tasks with high accuracy"
Postdoctoral researcher Dr Omid Rohanian says, “Our project focuses on developing compact and efficient models that can perform biomedical tasks with high accuracy. This approach involves a method known as knowledge distillation, which allows developing lightweight and efficient models from larger and more resource-intensive models with minimal performance loss”.
The pretrained models can be utilised in a wide range of biomedical tasks, such as named entity recognition, relation extraction, and question answering. For example, in named entity recognition, which involves identifying entities such as diseases, genes, and proteins in unstructured biomedical texts, these models can help researchers and healthcare professionals to identify and understand complex medical conditions more accurately and quickly. In addition, they can extract valuable information from biomedical texts by identifying relationships between different entities. This is particularly useful for unlocking important insights from large datasets, and has the potential to improve clinical decision-making.
The research, "On the Effectiveness of Compact Biomedical Transformers", has recently been published in the Oxford Bioinformatics journal, the official journal of the International Society for Computational Biology (ISCB).




